Research
Most life on Earth is microbial. Animals depend on complex ecosystems of microbes that live in and on us: microbiomes. The dense and diverse microbiome found in our gut plays essential roles in digestion, immunity, and many other aspects of vertebrate physiology. We are interested in developing a more detailed description of the roles of the individual members of this ecosystem and how they vary across host species, lifestyle, and health and disease.
Our research projects use both computational and experimental tools and approaches to varying degrees.
Comparative genomics of human and animal gut microbes
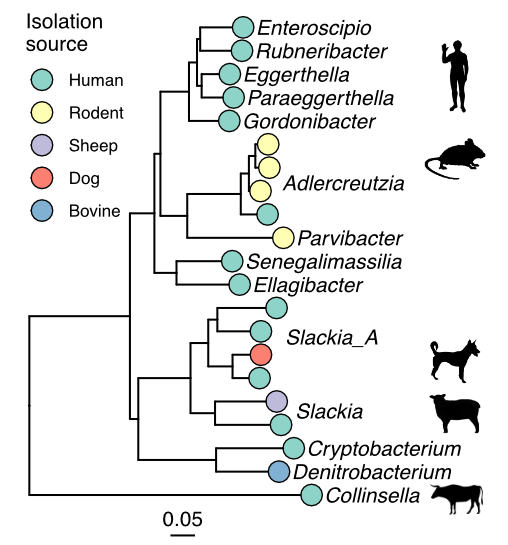
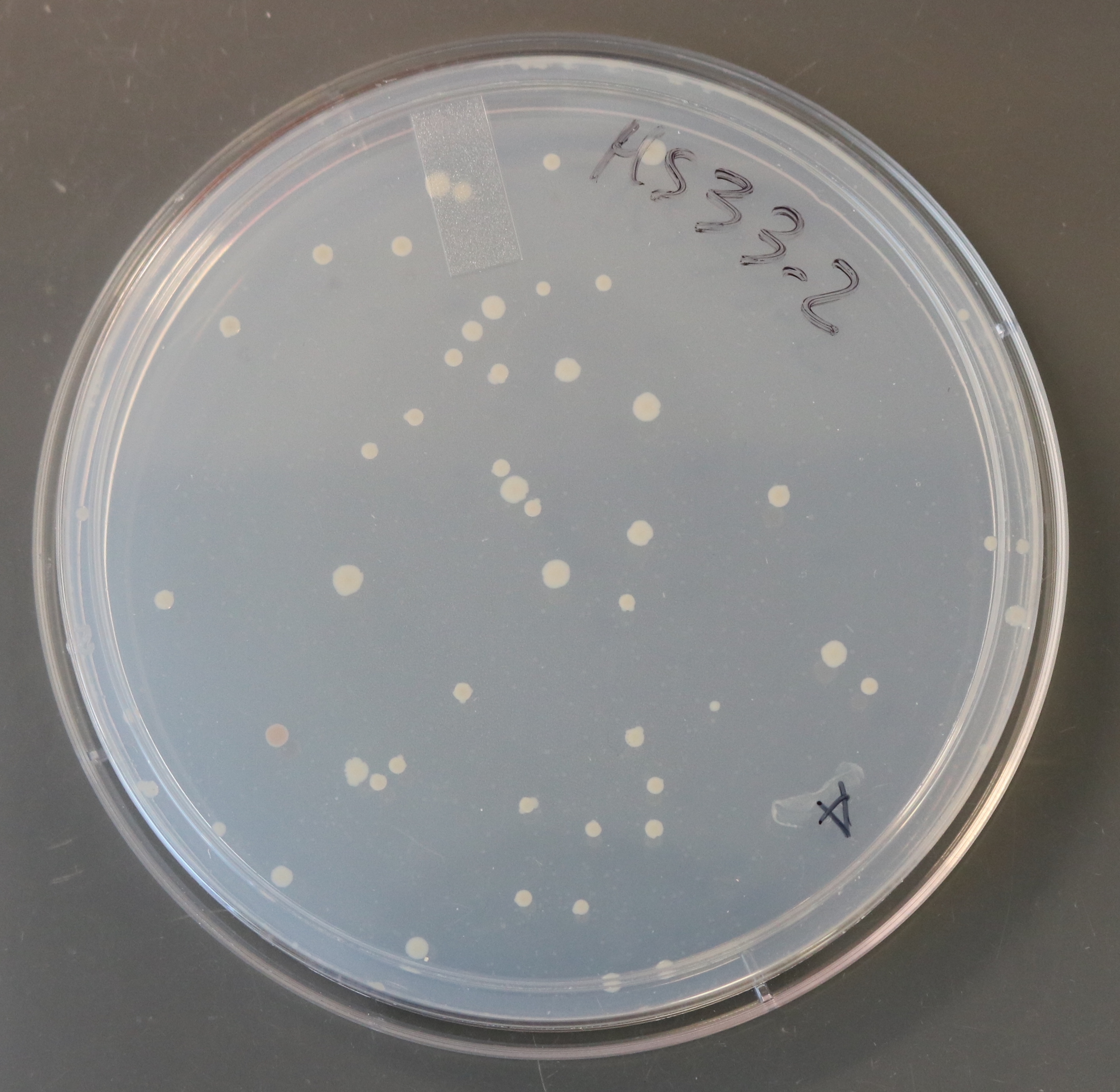
How do the resident microbes in your gut differ from their relatives living in the gut of your dog? Or from someone halfway across the world? How did their unique genetic and metabolic capacities evolve? What effects might they have on the physiology and ecology of the gut? We use computational genomics and experimental characterization of new isolates to explore these questions, with a focus on the prevalent and metabolically diverse bacterial family Eggerthellaceae.
Metabolic strategies of human gut bacteria
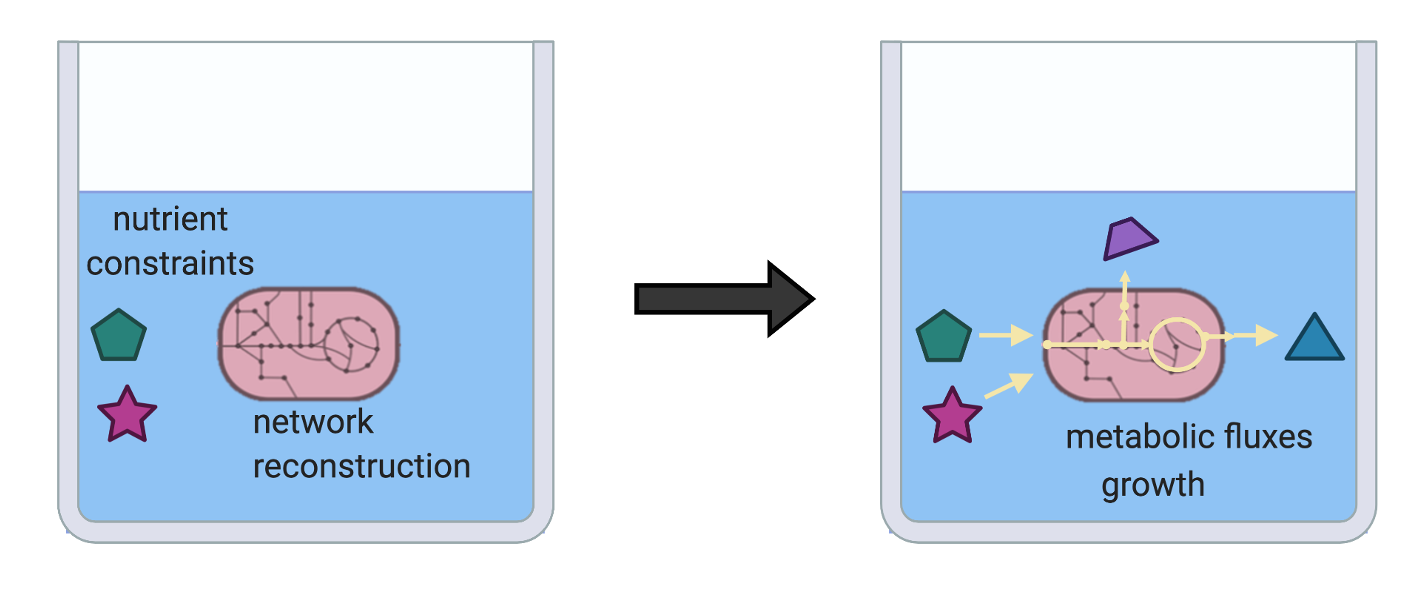
The most abundant microbes in your gut break down dietary fibers into smaller compounds called short-chain fatty acids. However, other gut microbes may rely on protein, lipids, or other nutrients from the diet or provided by the host. How do these nutrients support the growth of these microbes? Can we use computational modeling to analyze their growth and predict their interactions with other community members?
Meta-omic microbiome analysis
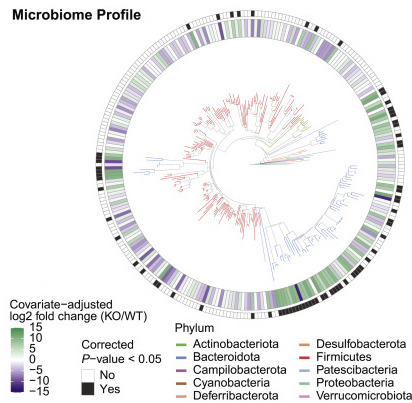
We enjoy collaborating with other researchers on experimental design, methodology, data processing, statistical analysis and/or data visualization for microbiome studies. Past projects have explored interactions between host genetics and the microbiome, microbiome sharing between humans and animals, novel microbial diversity in wild animals, and other topics, using 16S rRNA amplicon sequencing, metagenomics, and metabolomics. If you have a collaboration idea, please feel free to get in touch.